Research in the lab is broadly focused on studying the processes

of, and mechanisms underlying adaptation, evolutionary innovation, and phenotypic diversity using comparative genomic, functional genomic, and population genomic approaches. Our research involves both the generation and analysis of diverse ‘omic’-scale data, including genomic, transcriptomic, phylogenomic, and epigenomic data. Our research is primarily motivated by broad biological questions and driven by empirical data, and this motivation often leads to the development of new data generation techniques (often leveraging next-generation sequencing), development of new theory, and development of new computational approaches that have broad relevance beyond our own research. We are not wedded to a particular model system, although we often focus on snakes as models because they provide an excellent system for studying extremes of adaptation and evolutionary innovation. We also value the development of effective and synergistic collaborations, and this has led to our involvement in a diversity of projects that address basic and medically relevant research questions that are related to adaptation, evolution, genomics, and computational biology.
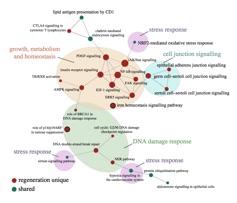
Comparative Mechanisms and Functional Genomics of Regenerative Organ Growth
Some snakes have evolved the ability to drastically and reversibly remodel their organs, metabolism, and physiology upon feeding. In pythons for example, the heart, intestine, liver and kidneys undergo unprecedented regenerative organ growth (enlarging 40-150% in 48 hours) after fasted animals are fed a large meal. Our work aims to understand the molecular and cellular mechanisms that underlie these reversible growth episodes, and to test the if such mechanisms are conserved across vertebrates, including humans – such knowledge would hold great translational potential for directing organ regeneration for treatment of human diseases. One key feature of snakes is that different lineages experience very different regenerative growth phenotypes (and some experience none at all), indicating snakes hold great comparative potential for dissection of mechanisms that direct regenerative growth. Together with several collaborators, we are integrating transcriptomic, proteomic, histological, tissue culture, and (most recently) mammalian organoid system approaches to develop and test hypotheses for the mechanisms underlying these growth responses in snakes, and the degree to which these mechanisms are conserved across vertebrates. To date, our work has provided exciting results indicating that the snake organ regeneration model has diverse translational potential for providing novel insight into the breadth of function of key vertebrate signaling pathways, and for the treatment of a spectrum of human diseases.
Adaptive Systems Genomics using Reptiles as Models
We are using snake and other reptile genomes to test hypotheses regarding
the evolution of complex traits, such as the evolution of vision, the evolution of extreme metabolism, and the evolution of highly toxic venom systems. Snake also provide an excellent example for studying the impacts and drivers of large-scale molecular convergent evolution. Our previous work has provided unique insight into unprecedented levels of genome-wide adaptation in snakes, the largest known example of convergent molecular evolution, the evolution of venom systems, and the remarkably slow evolution of crocodilian genomes. Ongoing projects target questions related to the evolution of vision, the role of genome structure in driving venom systems, the loss of limbs, evolution of color pattern and mimicry systems, evolution of toxin-resistance, and the adaptive co-option of conserved organ growth pathways. We have recently expanded to incorporate population-level genomic sampling into many of these questions. This work is important for understanding mechanisms underlying adaptation, evolution of complex systems and phenotypes, and the non-canonical function of important pathways involved in human development and disease.
Population Genomics of Adaptation and Speciation
Driven by our interest in adaptation, we have initiated several empirically-driven studies that use snakes as model systems to investigate adaptation at the population level in both natural and introduced populations. These studies are driven by fundamental questions including the roles of divergence and gene flow in adaptation and speciation, the roles of adaptation in species invasions, and the degree to which convergent phenotypic outcomes are driven by convergent molecular changes in different populations and lineages. We have been funded by NSF (through two DDIG grants) to initiate two studies – one focused on the molecular basis of convergent island dwarfism phenotypes in Boa constrictors, and a second that focuses on shared and unique adaptations in populations of rattlesnakes, and how these adaptive loci promote speciation. We have also conducted substantial population genomic work on introduced populations of Burmese Pythons in Florida, in which we are finding evidence for in-situ adaptation.
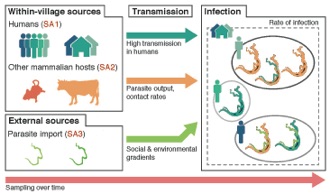
Population Genetics, Epidemiology, and Genotype-Pathogenesis Analysis of Parasites
We are currently applying approaches that what we know well from other projects – analysis of
genome-wide SNPs to understanding population genetic structure and genotype-phenotype associations – to understanding the population genetic structure, epidemiology of re-infection, and parasite genotype-patient outcomes for
Schistosoma (blood fluke) parasites that cause shistosomiasis in Asia. Working with collaborators studying
Schistosoma epidemiology in China, we have developed approaches to collect population genomic-scale data from current and historical shistosome samples to provide retrospective and prospective analyses of outbreak dynamics, basic yet currently not known information on infection biology, and reinfection information. Our goals are to use these data to better understand and thus control shistosome infection, and to use this genomic sampling to detect the evolution of resistance to drugs used to treat shistosomiasis (which would be catastrophic if not adequately prepared for).
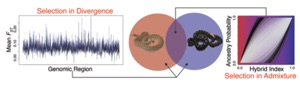
Theoretical Phylogenomics & Species Delimitation with Genome-Scale Datasets
Ironically, massive datasets have made inferences of phylogeny and
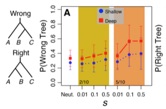
species limits more difficult in many ways – a major problem is that seemingly small systematic biases in existing methods are compounded by large datasets, leading to surprisingly high rates of error. This is an urgent issue because inferences of phylogeny and the delimitation of species are essential components of modern biology and for the fields of evolutionary biology and comparative genomics. For example, we previously discovered that traditional likelihood (and Bayesian) methods of phylogenetic inference can be grossly inaccurate when adaptive and convergent events occur. Similarly, our recent work on species delimitation indicates that these inferences can also be grossly mislead by small amounts of admixture and selection. We are developing new approaches for phylogenomic and species delimitation inference that better account for natural complexities of speciation, including the processes of admixture and selection.
Dissecting the Links Between Genome Structure, Function, and Evolution
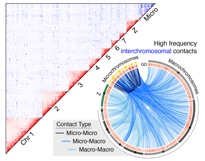
We are interested in the fundamental interactions of genome structure, genome function, and genome evolution. Presently we know that aspects of genome structure (e.g., sex chromosome differentiation, chromatin structure, transposable elements (TE), and GC isochore structure) are in some way linked to aspects of genome function (e.g., recombination, TE silencing, gene conversion) and genome evolution (e.g., insertion and deletion, mutation). The precise cause-effect relationships among these features, however, remain relatively poorly known. Our past and ongoing research indicates that various snake lineages show extraordinary shifts in many aspects of genome structure, function, and evolution, making them excellent model systems. To enable this work we have developed new methods for analysis of genomic TEs, for surveying TEs in unsequenced genomes, and for testing hypotheses for TE impacts on recombination. Our results suggest that REs have contributed in key ways to changes in multiple aspects of genome structure and function, and that these changes in genome structure have contributed to the evolution of extreme phenotypes including the evolution of venoms.
Omic Mechanisms Underlying the Evolution of Transgenerational Plasticity
Environmentally-driven changes in phenotypes that span multiple generations have been an area of interest and contention since Lamarck. Such ‘transgenerational plasticity’ (TGP) occurs when the environment experienced by parents alters the
phenotypes of subsequent generations. Although TGP appears to be widespread, our understanding of how and why it works is poor. Working with collaborators, we are using
Daphnia (water fleas) as a model system to test hypotheses for the conditions that drive the evolution of TGP in populations, and the hierarchical mechanisms by which TGP evolves, occurs, and manifests in eco-responsive phenotype changes. In addition to genomic and transcriptomic approaches, we have developed multiple new approaches for economically surveying the epigenome of samples for this project that are broadly applicable to other systems.
Todd Castoe’s Home Page:
http://www.snakegenomics.ToddCastoe/Home.html